

GBS-Div is provided "as-is" and without warranty of any kind, express, implied or otherwise, including without limitation, any warranty of merchantability or fitness for a particular purpose. In no event shall the authors, the CIRAD or its departments be liable for any special, incidental, indirect or consequential damages of any kind, or any damages whatsoever resulting from loss of use, data or profits, whether or not advised of the possibility of damage, and on any theory of liability, arising out of or in connection with the use or performance of this software.
GBS-Div is compatible with all current Microsoft Windows operating systems
GBS-Div is developed with Microsoft Windows Visual Basic Studio.Net targeted on Microsoft .Net Framework 4.0.
The use of GBS-Div under Windows emulators for MacOS or Linux has been reported by several users but we have not tested these compatibilities.
GBS-Div has no internal limitations in data size other than RAM memory and hard disk space availability. Analyses of large datasets can be time consuming, depending on the frequency and the core number of the processor.
GBS-Div is not open source but it is provided free for use in research and education. The download package includes the Software installation.
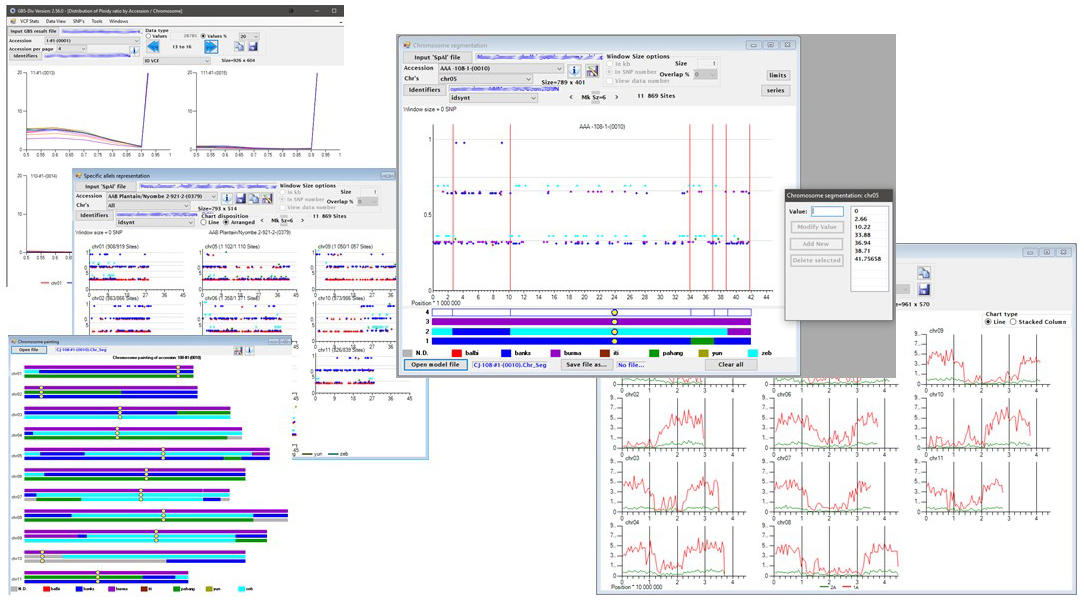
GBS-Div explores VCF files of SNP polymorphisms observed on a set of accessions. It proposes a series of filters to clean these data as well as statistical indicators to evaluate the effect of these filters. The ploidy ratio is used to estimate the local ploidy of each accession. Mixtures of ploidy levels between accessions, between chromosomes of the same accession or even within the same chromosome (duplications) are accepted and a file of the ploidies actually observed allows a genotype calling based on the actual local ploidies. Dissimilarity matrices can be computed on the whole genome, on a particular chromosome or even on a segment of this chromosome. The resulting .DIS files can be used directly in DARwin. If ancestral poles are present in the set of accessions, the specific loci of these ancestors can be identified and their occurrence in particular accessions can then be tracked. Interactive tools are available to explore the mosaic of ancestor contributions along the chromosomes and to establish, store and represent the chromosome paintings.
GBS-Div is designed to work with ploidy levels between x and 4x exclusively.
If by accident you notice that GBS-Div has not been updated correctly, uninstall the local version before reinstalling the new one.